David Odekunle
What is CRISPR?
CRISPR, which is short for clustered regularly interspaced short palindromic repeats, allows for genomic editing. It is derived from the natural defense mechanism of bacteria to protect against invading bacteriophages, which are viruses that infect bacteria and can reproduce inside them.
How does it work?
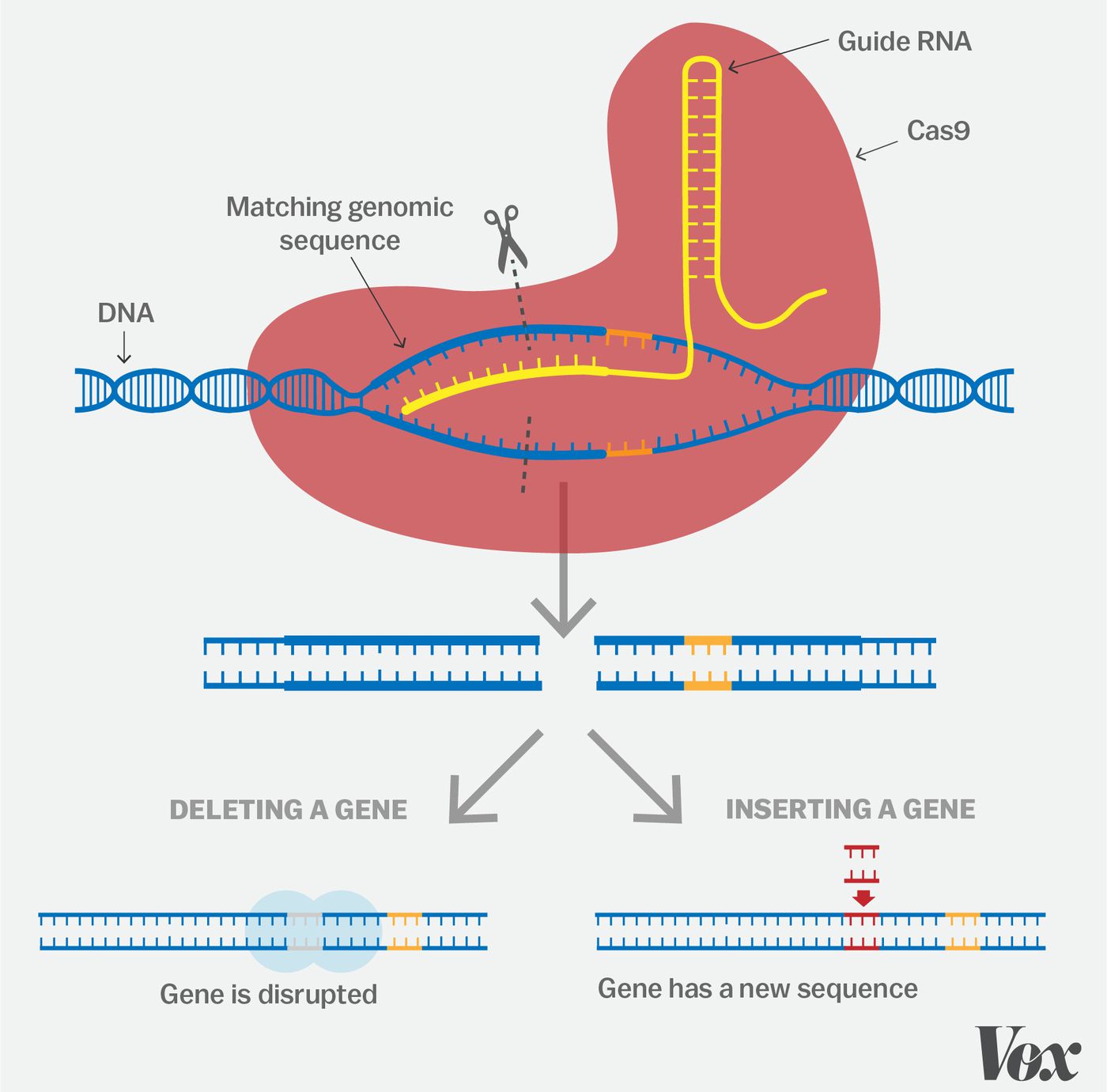
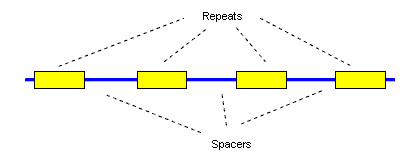
CRISPR genome editing has two vital phases: immunization and immunity. Before we explore these processes, let us familiarize ourselves with the terms involved. We have spacer sequences which are short segments that are identical to the invading bacteriophage DNA. Additionally, we have repeat sequences, which exist in the CRISPR array and are separated by spacer sequences. The pattern is typically repeat, spacer, repeat, spacer, and so on. There are various Cas proteins, but for our simplicity, we will focus on three main types: Cas1, Cas2, and Cas9.
During the immunization phase, the spacers are acquired. We initially begin with a CRISPR array complex with repeat sequences and gaps in between. When an invading bacteriophage enters the bacteria, Cas1 and Cas2 complexes recognize the Protospacer Adjacent Motif (PAM) site on the bacteriophage. The Cas proteins bind to an area adjacent to the PAM site, cleaving part of the bacteriophage DNA and inserting it into the CRISPR array. This step is essential because it is the spacer sequence that will help with later recognition of reinfecting bacteriophage.
Following immunization, we have the immunity phase, where the invading DNA from the bacteriophage is destroyed upon reinfection. First, upon bacteriophage reinfection, we have the CRISPR array, which now includes the repeats and spacers being transcribed to RNA. Then, tracrRNA, which has a sequence complementary to the CRISPR repeat sequence can bind and stabilize the CRISPR RNA. This stabilization allows for Cas9 protein to bind to the CRISPR RNA and tracrRNA. While there are different processing mechanisms, we will use a typical method where RNase digests the complex made up of CRISPR RNA, tracrRNAs, and Cas9 proteins. This leaves individual complexes, each containing one spacer sequence, cas9, tracrRNA, and CRISPR RNA. Now, when the bacteriophage reinfects, the spacer sequence recognizes a sequence complementary to the phage, and the cas9 recognizes to and makes a double stranded cut at a sequence adjacent to the PAM site.
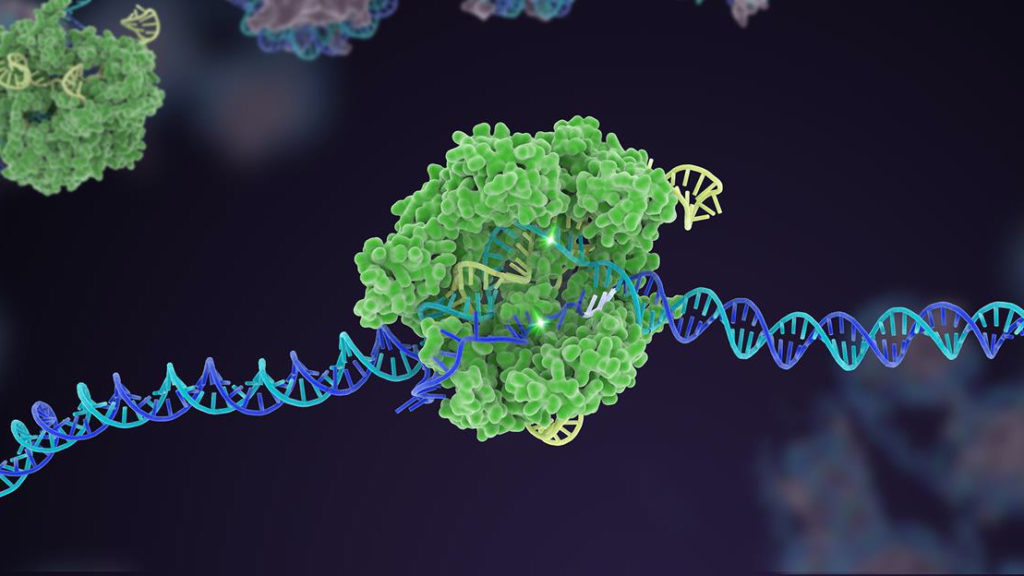
Why are these double stranded breaks important for gene editing?
Double stranded breaks in DNA are lethal and highly toxic if they are not fixed. There are two main methods of double stranded break repair: Non-homologous end joining (NHEJ) and Homology Dependent Repair (HDR). NHEJ typically results in frameshift mutations which are insertions or deletions that occur in the DNA strand. Let’s take a strand reading 5’ A T G C 3’. If it experiences an insertion, the new strand could read 5’ A T A G C 3’. The insertion here is the adenine nucleotide. Because of NHEJ typically resulting in random mutations, we can randomly manipulate a target sequence. Typically for this repair, the broken ends are bound to by Ku protein complex. Then the ends of the break are trimmed by nucleases, and the break is sealed by ligase. The second form of repair, which is far more complex, but also more effective in creating a desired mutation is HDR. Homology Dependent Repair results in point mutations, which are mutations where a single nucleotide is changed, which results in an amino acid change. Let’s take a strand reading 5’ A C G T 3’. If it experiences a point mutation, the new strand could read 5’ A G G T 3’ where the cytosine is altered to guanine. For homology-dependent repair, we need a homologous sequence that through DNA synthesis will help fill in the missing DNA on the target strand. Both techniques are essential to editing DNA in such a way that different genes can be targeted, which will lead to amino acid changes, and thus render changes in genotype and phenotype.
Concluding Thoughts
CRISPR gene editing involves the CRISPR-Cas9 creating a double-strand break in DNA and the repair of the break results in frameshift mutations or directly point mutations that may foster certain changes in protein function and thus affecting the genotype and perceived phenotype.